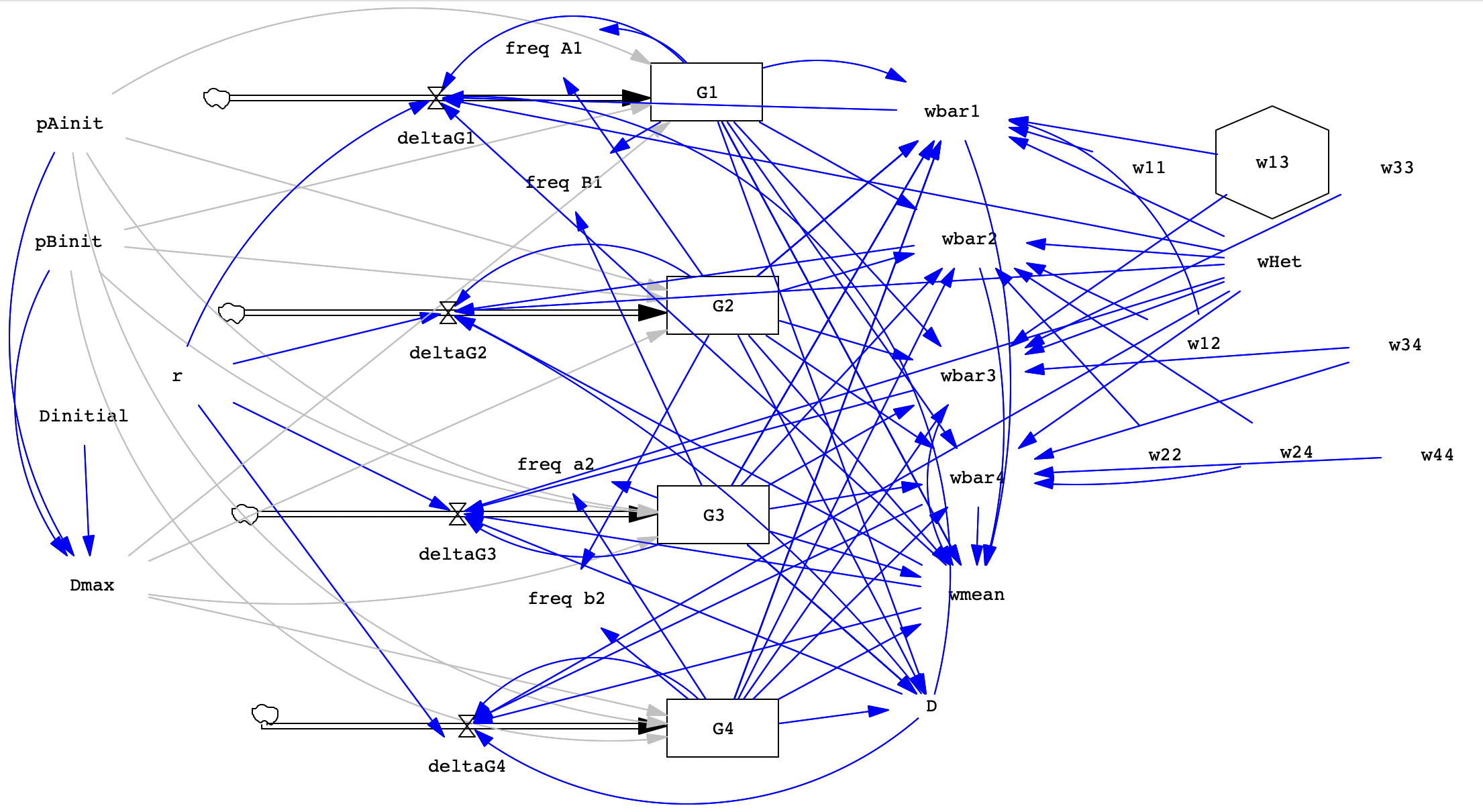
Overview
Today we’ll build a two-locus model to allow us to explore the phenomenon of gametic disequilibrium, including how it is affected by physical linkage and by natural selection. Just like we did with drift, we’ll create a toggle to allow us to start out either in maximum Disequilibrium or not (that is, in equilibrium). Until now, we’ve tracked genotypes and also allele frequencies. However, the two-locus model has 9 possible genotypes so we are only going to track the four possible gamete types with our stocks. We will account for genotypes when modeling selection by specifying fitness values for each of the 9 genotypes. The inclusion of a spinner for r, the rate of recombination, will allow us to explore the effects of physical linkage. These parameters will then let us see how it’s possible to have gametic disequilibrium even when the loci are not physically linked (r = 0.5).
Because we are tracking gametotypes rather than genotypes in this model, we’ll have to start from scratch, though you should notice similarities to the models we built in earlier lab exercises.
Before we get started, let’s think about what it means for the population to be in maximum disequilibrium for the alleles at two loci (Alpha and Beta loci). We have two alleles for each locus with the following allele frequencies:
| Locus Alpha, alleles A and a | Locus Beta, alleles B and b |
| freq A = frequency of the A allele | freq B = frequency of the B allele |
| freq a = frequency of the a allele | freq b = frequency of the b allele |
Now, if we assume that loci are independently assorting, then we know the probability of having each combination of alleles in a gamete – it’s just the product of the two allele frequencies.
G1 = frequency of the AB gamete = freq A * freq B
G2 = frequency of the Ab gamete = freq A * freq b
G3 = frequency of the aB gamete = freq a * freq B
G4 = frequency of the ab gamete = freq a * freq b
If we imagine a population where freq A = 0.5 and freq B = 0.5, then we expect G1 = 0.5*0.5 = 0.25. And we’d also end up with 0.25 for the frequencies of G2, G3, and G4. That would be our prediction if the Alpha and Beta loci assort independently.
But now imagine that when we look at gametes, we see that what we have is:
G1 = AB gamete = 0.5
G2 = Ab gamete = 0
G3 = aB gamete = 0
G4 = ab gamete = 0.5
We still have all the allele frequencies equal to 0.5 BUT the way alleles are paired in gametes is not random - we only get AB and ab. This would be maximum disequilibrium. The actual maximum disequilibrium values depends on the allele frequencies since you can’t have more AB gametes than you have A alleles, for example. Below, I give you the equations that will help us figure out the maximum for whatever starting conditions we choose.
In our model, we will set it up so we can choose to start with maximum disequilibrium OR start in gametic equilibrium.
Here's a reference image of the completed model: