Flowering Plant Phylogenomics
| Above: A circle tree that depicts the relationships among the major lineages of flowering plants as determined from plastid genome sequence data. Starting at the bottom center (at the base of the tree) and proceeding counterclockwise, are images of Amborella, Nymphaea, Illicium, Chloranthus, Piper, Liriodendron, Ceratophyllum, Ranunculus, Pelargonium, Helianthus, Yucca, Triticum, and Acorus. The background picture is of the island of New Caledonia, the home of Amborella. To see the paper associated with this image, click here. Click on the image to see a larger version. |
Plants contain genetic material in three separate cellular compartments: the nucleus, the mitochondrion, and the plastid (which, when used for photosynthesis—as it mostly is—is called a chloroplast). It may seem strange that two organelles should have their own genomes. Why do mitochondria and plastids have DNA? The circular nature of the both genomes reveals the answer: mitochondria and plastids represent highly reduced versions of what at one time (~2 billion years ago!) were free-living bacteria. As these ancestral bacteria took up residence within the eukaryotic cell and transitioned to fully integrated organelles, most of their genes were either lost or transferred to the host cell’s nucleus, leaving only several dozen genes that are most important to the day-to-day functioning of these organelles. As a result, the plant mitochondrial and plastid genomes generally range from 150,000-500,000 base pairs in length, while a typical plant nuclear genome would range from 0.5-4.0 billion base pairs (bp) in length!
The two plant organellar genomes differ in several fundamental ways. Unlike the animal mitochondrial genome, which has an extremely compact organization, the plant mitochondrial genome has a more fluid organization due to the relatively high proportion of A/T-rich (as opposed to C/G-rich) intergenic spacer regions, which tend to accumulate repeat sequences. In addition, the plant mitochondrial genome has a relatively high propensity to incorporate foreign DNA, such that some mitochondrial genomes in plants have genes or entire regions that were acquired from other genomes at some point during their evolutionary history. As a result of these characteristics, plant mitochondrial genomes vary in size by roughly an order of magnitude across flowering plants (from <400,000 bp in some species to >10 million bp in Silene!). In stark contrast stands the chloroplast genome. The chloroplast genomes of almost all flowering plants share an identical gene content and arrangement, with much less intergenic spacer and, in general, few or no significant repeat sequences. Plastid genomes essentially never incorporate foreign DNA; despite active searching, almost no examples of horizontal gene transfer have been detected across all of plants (flowering or otherwise). Plastid genomes can vary in size, although nearly all flowering plant plastids have a genome from 150,000-170,000 bp in length.
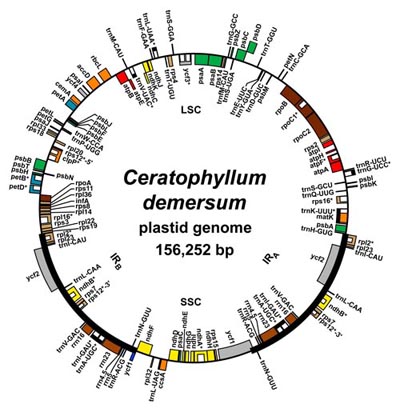 |
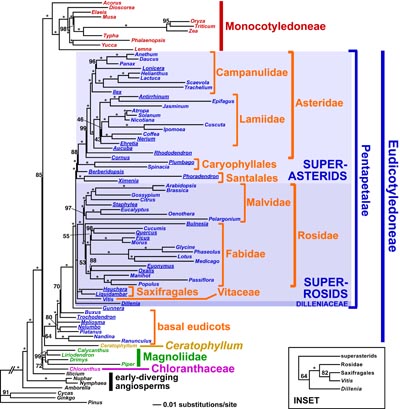
| Above: A map of a typical angiosperm plastid genome (click on the image for a larger version). Below: An overview of angiosperm relationships, taken from Moore et al. (2010), "Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots", PNAS. |
 |
Because of its structural stability and its uniparental inheritance (in other words, recombination among different copies of DNA does not occur, as it does in the nucleus, where there are maternal and parental copies of all genes), the plastid genome provides a critical source of information for reconstructing the evolutionary relationships of flowering plants. Indeed, the plastid genome has been the workhorse of plant systematics since the beginning of the molecular phylogenetic era. However, until recently most researchers relied on only a handful of genes to reconstruct plant phylogeny (phylogeny is the term we use to refer to the evolutionary relationships within a group). Recent advances in DNA sequencing technology over the past five years now enable the rapid and (relatively) cheap sequencing of entire plastid genomes, thus increasing the amount of available sequence data 10-50x over what has historically been used in plant phylogenetics.
Plant systematists have made great strides in reconstructing angiosperm (flowering plant) phylogeny over the past 20 years, mostly through the analysis of a handful of plastid genes. The overall backbone of angiosperm phylogeny is now well-established (see figure at right). This new view of angiosperm relationships has played a central role in our developing understanding of numerous aspects of angiosperm evolution, including the origins and evolution of the flower. However, resolving many relationships in the deeper levels of angiosperm phylogeny has been impossible using only a few genes.
Plastid genomics raises the possibility of finally resolving outstanding issues in the flowering plant Tree of Life. Theoretical as well as some experimental work has suggested that greatly increasing the number of base pairs in a phylogenetic analysis can resolve recalcitrant phylogenetic relationships under certain conditions. As part of the Angiosperm Tree of Life project, I helped sequence the complete plastid genomes of more than 30 flowering plants, representing most major angiosperm groups, in an effort to test this idea in flowering plants. This sequencing effort has allowed phylogenetic data sets up to almost 70,000 bp in length to be analyzed. Analyses of these data sets have provided the first reasonably strong support for a completely resolved tree of basal angiosperms, and have identified three well-supported major lineages (called superasterids, superrosids, and Dilleniaceae) that encompass nearly all eudicot angiosperms, which represent more than 70% of all flowering plants. Given the success of this approach, we are working with the Soltis lab to sequence additional strategically selected angiosperm plastid genomes to help resolve other outstanding issues in flowering plant phylogeny. Currently we are focusing on the large clade Caryophyllales, which comprises perhaps the most physiologically and ecologically diverse group of angiosperms: as a whole, the group has adapted to every terrestrial habitat type on earth, and includes such amazing plants as cacti, pitcher plants, and living stones.
I am also working with Dr. Stephen Smith of the University of Michigan and Dr. Sam Brockington of the University of Cambridge to leverage the latest DNA sequencing technology to sequence thousands of nuclear genes for Caryophyllales, both to resolve relationships within the clade and to explore whether rates of molecular evolution are correlated with changes in ecophysiology. We have just begun this project, which involves sequencing the transcriptomes (the portion of the genome that is actively transcribed) of numerous species in the clade.
Top
Last updated on December 31, 2012
All images are the copyright of Michael J. Moore
