Manual
Part B: Making our s more "subthalamic"
B.1.a Changing membrane properties
As mentioned in part A1, the built-in passive and active channel properties (pas and hh respectively) are not appropriate for subthalamic s. We can modify the passive property parameters in the dendrites and soma to reflect correctly the passive membrane properties of rat subthalamic nucleus projection s. To modify the active channel properties we will need to build new active channels (see part D) using the model description language (NMODL). For the moment we will simply modify the standard hh channel densities.
We recommend that you open CellGUI.hoc and save it as Thalamic1.hoc. As in previous tutorial, type the code that is highlighted into the file. Remember to save it periodically. Save it in the same folder as the others
Your Thalamic1.hoc file should look like the following:
load_file("nrngui.hoc")
ndend = 2
create soma, dend[ndend]
access soma
soma nseg=1
soma diam=18.8
soma L = 18.8
soma Ra=123.0
insert hh
insert pas
dend[0] { nseg = 5 diam = 3.18 L = 701.9 Ra = 123 insert pas }
dend[1] { nseg = 5 diam = 2.0 L = 549.1 Ra = 123 insert pas }
// Connect things together
connect dend[0](0), soma(0)
connect dend[1](0), soma(1)
objectvar electrode
soma electrode = new IClamp(0.5)
soma electrode.del=100
soma electrode.dur=100
soma electrode.amp = 0.45
print soma.v
forall psection()
tstop = 300
As described in part A1, we can access the passive and active properties once they are inserted in the section. For example, we can increase the soma sodium channel density by adding the code that specifies the hh properties:
insert hh
gnabar_hh= 0.25
insert pas
The properties of the passive membrane mechanisms in the soma and dendrites must also be changed. Subthalamic nucleus projection s in rat have a membrane time constant of 6msec. As we assume all cells have approximately the same membrane capacitance (1 µF/cm2), by using the assumption of an RC circuit we can calculate the subthalamic nucleus projection membrane resistance from:
time constant = Resistance * Capacitance
This yields a membrane resistance of 6000 ohms cm2. Remember, requires this as a conductance g_pas (which is the reciprocal of the resistance, i.e. 0.0001667 S/cm2). Subthalamic nucleus cells also have a very high resting membrane potential, so our leakage equilibrium potential (e_pas) is set to -60mV. The dendrite definition then becomes (for dendrite 0):
dend[0] { nseg = 5 diam = 3.18 L = 701.9 Ra = 123 insert pas g_pas = .0001667 e_pas = -60.0 }
Note, you must make the same passive changes in the other dendrite.
dend[1] { nseg = 5 diam = 2.0 L = 549.1 Ra = 123 insert pas g_pas = .0001667 e_pas = -60.0 }
The hh channels inserted into the soma also contains passive properties, called gl_hh and el_hh. These can be modified in a similar manner, making our soma definition:
gnabar_hh=0.25
gl_hh = .0001666
el_hh = -60.0
Save the file with the changes we suggested and run the file by finding it and double clicking on it.
Once it has been opened and you can access the Main Menu window, select Graph>>Voltage axis and then run the program again. You should see the following graph:
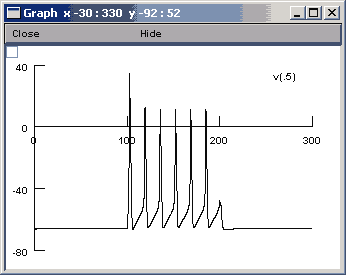
If you have problems running and visualizing the results go back to the "Using the GUI interface" tutorial.
B.1.b Avoiding tedious typing: for loops
We will modify our Thalamic1.hoc file dramatically through this process. Therefore it is advisable to make a working copy of Thalamic1.hoc. To do so "save as" the Thalamic1.hoc file as Thalamic2.hoc. We will work on Thalamic2.hoc.
In the previous section, you should have inserted passive channels and set their properties for dend[0] and dend[1] individually. This is OK when there are only two dendrites, but what if there were 100 dendrites that we wanted to insert passive conductances into? We'd have to type out 100 times three lines or do lots of cutting and pasting; boring and liable to lead to repetetive stress syndrome (RSI).
The solution is the for loop. A for loop can step through each element in an array in turn. The format of the for loop is:
for var = lower-bound, upper-bound command
where var is the loop variable, lower-bound is the lower bound
of the loop, upper-bound is the upper bound of the for loop and command is
the command we want to run through the loop. Thus to insert passive conductances
and change their properties in the dendrites we can replace the bold-green
text in the following statements
dend[0] { nseg = 5 diam = 2.0 L = 549.1 Ra = 123 insert
pas g_pas
= .0001667 e_pas = -60.0 }
dend[1] { nseg =
5 diam = 2.0 L = 549.1 Ra = 123 insert
pas g_pas = .0001667 e_pas = -60.0 }
with the following line
for i = 0, ndend-1 dend[i] { insert pas g_pas = .0001667 e_pas = -60.0 }
This statement doesn't replace the whole line which defines dend[1] and dend[0]. It only substitutes for the parts that are consistant across all dendrites (in bold-green).
Remember that arrays, of which the dendrites are defined, are indexed by starting with the number zero not 1. This is why the for loop begins with zero and goes to the number of dendrites minus 1.
Also, since the nseg and Ra for both dendrites is similar, we could have added them to the for loop. Since these are probably different in future segments we didn't add them here. If we had added them the code would look like this:
for i = 0, ndend-1 dend[i] { nseg = 5 Ra=123 insert pas g_pas = .0001667 e_pas = -60.0 }
To program for loops, we have had to make many changes. Run the simulation again and be sure it is working. If you get the same data/graphs as you saw in B.1.a, you're on the right track. If not, refer to Thalamic2.hoc in the code section of the tutorial folder.
The code in Thalamic2.hoc should look like this:
load_file("nrngui.hoc")
ndend = 2
create soma, dend[ndend]
access soma
//Define membrane mechanisms
soma nseg=1
soma diam=18.8
soma L = 18.8
soma Ra=123.0
insert hh
gnabar_hh= 0.2
gl_hh = .0001666
el_hh = -60.0
insert pa
dend[0] { nseg = 5 diam = 3.18 L = 701.9 Ra = 123}
dend[1] { nseg = 5 diam = 2.0 L = 549.1 Ra = 123 }
for i = 0, ndend-1 dend[i] { insert pas g_pas = .0001667 e_pas = -60.0 }
// Connect things together
connect dend[0](0), soma(0)
connect dend[1](0), soma(1)
// create an electrode in the soma
objectvar stim
stim = new IClamp(0.5)
stim.del = 100
stim.dur = 100
stim.amp = 0.45
print soma.v
forall psection()
tstop = 300
Note: since 'electrode' is simply the named we gave to the object we created in the previous tutorials, in practicality it can have any name. In this code, it is referred to as 'stim.'
Summary
- Changing the properties of the soma and dendrite to make them more "subthalamic"
- Visualize the results by running the program and opening a voltage graph
- Reducing the number of lines needed by implementing a for loop
Next: Creating a realistic dendritic tree
Acknowledgements
Last modified on 10/29/04 by
Albert Borroni, CS/Neuoriscience Oberlin College (aborroni@oberlin.edu)
based on tutorial written by
Andrew Gillies (andrew@anc.ed.ac.uk)
David Sterratt (dcs@anc.ed.ac.uk)
which is originally based on the tutorials by Kevin E. Martin
with the assistance of Ted Carnevale and Michael Hines
Acknowledgements
Last modified on 10/29/04 by
Albert Borroni, CS/Neuoriscience Oberlin College (aborroni@oberlin.edu)
based on tutorial written by
Andrew Gillies (andrew@anc.ed.ac.uk)
David Sterratt (dcs@anc.ed.ac.uk)
which is originally based on the tutorials by Kevin E. Martin
with the assistance of Ted Carnevale and Michael Hines