Manual
The easiest way to make the change is to modify the bold text and delete the red text in the following section of code:
objectvar stim[nSThcells]
for i = 0,
nSThcells-1 SThcells[i].soma {
stim[i] = new IClamp(0.5)
objectvar stim
i = 1
SThcells[i].soma {
stim[i] = new IClamp(0.5)
C.2. Connecting the s together
After completing exercise C.1. we now have a very nice set of subthalamic s but we have not yet connected them together yet . Currently, they are operating as four independent s, each with an electrode placed in their soma. We now need to connect them together.
The modern form of connecting s together is in an event based simulation model. Events are usually things like action potentials, as it is the events (rather than, for example, the specific voltage levels) that we need to pass or communicate between s. By using an event based model, we can dramatically reduce the amount of inter- communication. This is important in simulation time optimisation and in simulations on parallel machines.
In this section we will extend our previous model, SubThalN1.hoc. Therefore you should open a new text file called SubThalNucl2.hoc and copy the code into this new file. Call this new file SubThalNucl2.hoc. Alternatively you could open the supplied code SubThalNucl1.txt and save it as SubThalNucl2.hoc.
To connect these 4 s to each other we must first add an additional public object variable to the template. The new object variable will refer to a list that will hold an arbitrary number of NetCon objects. A NetCon object is an object associated with a particular source of events (usually the voltage in a segment of the soma, where a particular change in Vm (action potential) will produce the binary event). So, we now begin our subthalamic nucleus code by adjusting the template in the following manner:
begintemplate SThcell public soma, treeA, treeB, nclist create soma, treeA[1], treeB[1] objectvar f, nclist proc init() {local i, me, child1, child2
The only thing we have done is to declare a new variable, nclist as a public variable and create an object called nclist. Next we will need to define the nclist by using the List() function.
f = new File()
nclist = new List()
AList is an
object that holds a list of other objects, like an array. In this case, the
object it will hold will be a synapse. We will see later
how to add items to a list and change their values.
The advantage of a list (over an array) is that we don't have to specify
in advance how big it will grow.
C.2.a Activating only 1 with our point process
Let's now modify the code so only one is being stimulated by an electrode. We will choose SThcells[1] to have an electrode placed in the soma and for it to pass depolarizing current between 100 and 200 ms from the start of the simulation run. (The code for setting up the point process is toward the bottom of the file). Delete the second line and replace it with a i=1.
| original | new |
objectvar electrode[nSThcells]
for i = 0, nSThcells-1
SThcells[i].soma {
electrode[i] = new IClamp(0.5)
electrode[i].del = 100
electrode[i].dur = 100
electrode[i].amp = 0.45
} |
objectvar electrode[nSThcells]
i = 1
SThcells[i].soma {
electrode[i] = new IClamp(0.5)
electrode[i].del = 100
electrode[i].dur = 100
electrode[i].amp = 0.45
}
|
Now, from 100 to 200 ms, SThcells[1] will be generating action potentials.
C.2.b Creating a synapse
In our first connection example, we only want to connect SThcells[1] to SThcells[0] and observe the EPSPs (Excitatory Post Synaptic Potentials) at the soma of SThcells[ 0].
In order for SThcells[1] to activate SThcells[0] we must first create some synaptic objects. Like IClamp which was discussed in A.2, a synapse is simply an object that can be positioned anywhere on a . For example, at the end of our current code we can define a new array of objects for synapses that will appear in various cells.section. We can create 10 objects which will hold information about synaptic contacts by placing following code immediately after the last curly bracket used to define our IClamp in SThcells[1].soma:
electrode[i].dur = 100
electrode[i].amp = 0.45
} maxsyn = 10 objectvar syn[maxsyn]
We will use the built-in synaptic type ExpSyn, which is a synapse whose conductance instantaneously rises on receiving a spike and then exponentially decays. We position the synapse, when it is created, in the same manner to IClamp i.e. by referring to a section.segment of a where we wish the synapse to be located and create the synapse there.
The code below creates an ExpSyn synapse positioned at the proximal end of treeA dendritic branch 22 on subthalamic 0 (marked in the diagram below by the small blue circle). The information for this synapse is stored in the syn array object, index 0 (syn[0]) which was created in the previous line.
access SThcells[0].treeA[22]syn[0] = new ExpSyn(0)
Saving and running this new hoc file will allow you to see the synapse. To see it you will need to select Tools>> Point Processes>>Managers>>Point Group. Right click on the graph, select 3D rotate, use the mouse to then rotate the set of s so that you can see all 4. Currently you only see the IClamp(1) process. To see the ExpSyn, select ExpSyn from the Show All button. You should then see a red dot indicating where the synapse is located on the dendrite of SThcell[0].
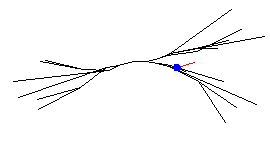
Note: in these displays, the red color shows which part is currently selected--click the mouse on the other object and that one will turn red.
C.2.c Using NetCon to connect and set activation trigger for synapse
Now we must define the source of events for this synapse. This will be the action potentials generated at the soma of SThcells[1]. Since SThcells[0] receives the events (i.e. this is the where the synapse is attached) we must append a new NetCon object to the nclist of this . To create a new NetCon object, we use the command format:
new NetCon(&source_v, synapse, threshold, delay, weight)
- source_v is the source voltage (in our case the voltage from SThcells[1].soma)
- synapse is the object variable that refers to the synaptic object receiving the events (in our case syn[0])
- threshold is the threshold which the voltage must reach for it to be considered that an action potential has occurred,
- delay is the connection delay
- weight is the connection weight strength at the synapse. The weight is a measurement of how strong the connection is: a perfect connection would have a weight of 1, a very weak connection might have a weight of 0.05.
So to connect SThcells[1] to the proximal end of treeA dendritic branch 22 on subthalamic SThcells[0] we add the command:
access SThcells[1].somaSThcells[0].nclist.append(new NetCon(&v(1), syn[0], 10, 1, 0.5))
First this command accesses SThcells[1].soma which is the area of our neuron that is being externally driven by the IClamp point process. Then the nclist of cell SThcells[0] has a new NetCon object appended. This NetCon object has a source voltage of SThcells[1].soma.v(1) (note, as we have accessed SThcells[1].soma in the command above, we need only use the short variable v(1)). The NetCon object applies to syn[0] which we have already attached to SThcells[0].treeA[22] (see above). Our threshold for action potentials is +10mV, our delay 1ms, and our synaptic weight 0.5.
Run the simulation and plot the voltage at SThcells[0].soma (or treeA[22]). You should see EPSPs resulting from the action potentials of SThcells[1]:
![SThcells[0].soma.v(0.5)](images/sthcells0.gif)
![SThcells[1].soma.v(0.5)](images/sthcells1.gif)
The first graph can be viewed by rightclicking on a voltage plot, then selecting "plot what." Click on "show" and select "all." From the first column, double-click on "SThcells[0]," from the second, "soma," and from the third, "v(.5)." Then run the simulation again.
C.2.D Working with lists
How do we change the weight, threshold and delay of our NetCon objects? If we had declared an objectvar called nc, we would have been able to refer to nc.weight, nc.threshold and nc.delay. However, our NetCon objects are embedded in a list, so we can't.
The solution is a group of commands into the terminal window that deal with lists. As mentioned above, the command
SThcells[0].nclist.append(obj)
appends(adds) the object specified by the object variable obj to the list nclist. We can use the command
SThcells[0].nclist.count()
to return the number of items in the list nclist. The command
SThcells[0].nclist.object(i)
returns the object at index i in the list nclist.
We can put these commands together to change properties of the connections. For example:
for i = 0, SThcells[0].nclist.count()-1 { SThcells[0].nclist.object(i).weight = 0.6 }
will change all of the weights onto SThcells[0] to 0.6.
Try playing with the connection parameters (delay, threshold and weight) and then connect more of the s together using more of the synaptic objects (but remember we have only created 10 objectvars to refer to synaptic objects in this example; feel free to add more by increasing the maxsyn parameter).
SUMMARY
- Activating only one rather then all 4 with our point process (electrode)
- Creating a synapse
- Using NetCon to connect s
- Working with lists
Next: Viewing Multiple s
Acknowledgements
Last modified on 10/29/04 by
Albert Borroni, CS/Neuoriscience Oberlin College (aborroni@oberlin.edu)
based on tutorial written by
Andrew Gillies (andrew@anc.ed.ac.uk)
David Sterratt (dcs@anc.ed.ac.uk)
which is originally based on the tutorials by Kevin E. Martin
with the assistance of Ted Carnevale and Michael Hines
Acknowledgements
Last modified on 10/29/04 by
Albert Borroni, CS/Neuoriscience Oberlin College (aborroni@oberlin.edu)
based on tutorial written by
Andrew Gillies (andrew@anc.ed.ac.uk)
David Sterratt (dcs@anc.ed.ac.uk)
which is originally based on the tutorials by Kevin E. Martin
with the assistance of Ted Carnevale and Michael Hines